Read in catalog information from a text file and plot some parameters#
Learning Goals#
Read an ASCII file using
astropy.ioConvert between representations of coordinate components using
astropy.coordinates(hours to degrees)Make a spherical projection sky plot using
matplotlib
Keywords#
file input/output, coordinates, tables, units, scatter plots, matplotlib
Summary#
This tutorial demonstrates the use of astropy.io.ascii for reading ASCII data, astropy.coordinates and astropy.units for converting RA (as a sexagesimal angle) to decimal degrees, and matplotlib for making a color-magnitude diagram and on-sky locations in a Mollweide projection.
import numpy as np
# Set up matplotlib
import matplotlib.pyplot as plt
%matplotlib inline
Astropy provides functionality for reading in and manipulating tabular
data through the astropy.table subpackage. An additional set of
tools for reading and writing ASCII data are provided with the
astropy.io.ascii subpackage, but fundamentally use the classes and
methods implemented in astropy.table.
We’ll start by importing the ascii subpackage:
from astropy.io import ascii
For many cases, it is sufficient to use the ascii.read('filename')
function as a black box for reading data from table-formatted text
files. By default, this function will try to figure out how your
data is formatted/delimited (by default, guess=True). For example,
if your data are:
# name,ra,dec
BLG100,17:51:00.0,-29:59:48
BLG101,17:53:40.2,-29:49:52
BLG102,17:56:20.2,-29:30:51
BLG103,17:56:20.2,-30:06:22
...
(see simple_table.csv)
ascii.read() will return a Table object:
tbl = ascii.read("simple_table.csv")
tbl
| name | ra | dec |
|---|---|---|
| str6 | str10 | str9 |
| BLG100 | 17:51:00.0 | -29:59:48 |
| BLG101 | 17:53:40.2 | -29:49:52 |
| BLG102 | 17:56:20.2 | -29:30:51 |
| BLG103 | 17:56:20.2 | -30:06:22 |
The header names are automatically parsed from the top of the file, and the delimiter is inferred from the rest of the file – awesome! We can access the columns directly from their names as ‘keys’ of the table object:
tbl["ra"]
| 17:51:00.0 |
| 17:53:40.2 |
| 17:56:20.2 |
| 17:56:20.2 |
If we want to then convert the first RA (as a sexagesimal angle) to
decimal degrees, for example, we can pluck out the first (0th) item in
the column and use the coordinates subpackage to parse the string:
import astropy.coordinates as coord
import astropy.units as u
first_row = tbl[0] # get the first (0th) row
ra = coord.Angle(first_row["ra"], unit=u.hour) # create an Angle object
ra.degree # convert to degrees
np.float64(267.75)
Now let’s look at a case where this breaks, and we have to specify some
more options to the read() function. Our data may look a bit messier::
,,,,2MASS Photometry,,,,,,WISE Photometry,,,,,,,,Spectra,,,,Astrometry,,,,,,,,,,,
Name,Designation,RA,Dec,Jmag,J_unc,Hmag,H_unc,Kmag,K_unc,W1,W1_unc,W2,W2_unc,W3,W3_unc,W4,W4_unc,Spectral Type,Spectra (FITS),Opt Spec Refs,NIR Spec Refs,pm_ra (mas),pm_ra_unc,pm_dec (mas),pm_dec_unc,pi (mas),pi_unc,radial velocity (km/s),rv_unc,Astrometry Refs,Discovery Refs,Group/Age,Note
,00 04 02.84 -64 10 35.6,1.01201,-64.18,15.79,0.07,14.83,0.07,14.01,0.05,13.37,0.03,12.94,0.03,12.18,0.24,9.16,null,L1γ,,Kirkpatrick et al. 2010,,,,,,,,,,,Kirkpatrick et al. 2010,,
PC 0025+04,00 27 41.97 +05 03 41.7,6.92489,5.06,16.19,0.09,15.29,0.10,14.96,0.12,14.62,0.04,14.14,0.05,12.24,null,8.89,null,M9.5β,,Mould et al. 1994,,0.0105,0.0004,-0.0008,0.0003,,,,,Faherty et al. 2009,Schneider et al. 1991,,,00 32 55.84 -44 05 05.8,8.23267,-44.08,14.78,0.04,13.86,0.03,13.27,0.04,12.82,0.03,12.49,0.03,11.73,0.19,9.29,null,L0γ,,Cruz et al. 2009,,0.1178,0.0043,-0.0916,0.0043,38.4,4.8,,,Faherty et al. 2012,Reid et al. 2008,,
...
(see Young-Objects-Compilation.csv)
If we try to just use ascii.read() on this data, it fails to parse the names out and the column names become col followed by the number of the column:
tbl = ascii.read("Young-Objects-Compilation.csv")
tbl.colnames
['col1',
'col2',
'col3',
'col4',
'col5',
'col6',
'col7',
'col8',
'col9',
'col10',
'col11',
'col12',
'col13',
'col14',
'col15',
'col16',
'col17',
'col18',
'col19',
'col20',
'col21',
'col22',
'col23',
'col24',
'col25',
'col26',
'col27',
'col28',
'col29',
'col30',
'col31',
'col32',
'col33',
'col34']
What happened? The column names are just col1, col2, etc., the
default names if ascii.read() is unable to parse out column
names. We know it failed to read the column names, but also notice
that the first row of data are strings – something else went wrong!
tbl[0]
| col1 | col2 | col3 | col4 | col5 | col6 | col7 | col8 | col9 | col10 | col11 | col12 | col13 | col14 | col15 | col16 | col17 | col18 | col19 | col20 | col21 | col22 | col23 | col24 | col25 | col26 | col27 | col28 | col29 | col30 | col31 | col32 | col33 | col34 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str24 | str25 | str9 | str6 | str16 | str5 | str5 | str5 | str5 | str5 | str15 | str6 | str5 | str6 | str5 | str6 | str4 | str6 | str13 | str14 | str26 | str23 | str11 | str9 | str12 | str10 | str8 | str6 | str22 | str6 | str19 | str23 | str9 | str38 |
| -- | -- | -- | -- | 2MASS Photometry | -- | -- | -- | -- | -- | WISE Photometry | -- | -- | -- | -- | -- | -- | -- | Spectra | -- | -- | -- | Astrometry | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- |
A few things are causing problems here. First, there are two header
lines in the file and the header lines are not denoted by comment
characters. The first line is actually some meta data that we don’t
care about, so we want to skip it. We can get around this problem by
specifying the header_start keyword to the ascii.read() function.
This keyword argument specifies the index of the row in the text file
to read the column names from:
tbl = ascii.read("Young-Objects-Compilation.csv", header_start=1)
tbl.colnames
['Name',
'Designation',
'RA',
'Dec',
'Jmag',
'J_unc',
'Hmag',
'H_unc',
'Kmag',
'K_unc',
'W1',
'W1_unc',
'W2',
'W2_unc',
'W3',
'W3_unc',
'W4',
'W4_unc',
'Spectral Type',
'Spectra (FITS)',
'Opt Spec Refs',
'NIR Spec Refs',
'pm_ra (mas)',
'pm_ra_unc',
'pm_dec (mas)',
'pm_dec_unc',
'pi (mas)',
'pi_unc',
'radial velocity (km/s)',
'rv_unc',
'Astrometry Refs',
'Discovery Refs',
'Group/Age',
'Note']
Great! Now the columns have the correct names, but there is still a
problem: all of the columns have string data types, and the column
names are still included as a row in the table. This is because by
default the data are assumed to start on the second row (index=1).
We can specify data_start=2 to tell the reader that the data in
this file actually start on the 3rd (index=2) row:
tbl = ascii.read("Young-Objects-Compilation.csv", header_start=1, data_start=2)
Some of the columns have missing data, for example, some of the RA values are missing (denoted by – when printed):
print(tbl["RA"])
RA
---------
1.01201
6.92489
8.23267
9.42942
11.33929
--
--
--
21.19163
21.5275
...
--
303.46467
321.71
--
--
332.05679
333.43715
342.47273
--
350.72079
Length = 64 rows
This is called a Masked column because some missing values are
masked out upon display. If we want to use this numeric data, we have
to tell astropy what to fill the missing values with. We can do this
with the .filled() method. For example, to fill all of the missing
values with NaN’s:
tbl["RA"].filled(np.nan)
| 1.01201 |
| 6.92489 |
| 8.23267 |
| 9.42942 |
| 11.33929 |
| nan |
| nan |
| nan |
| 21.19163 |
| 21.5275 |
| 25.49263 |
| nan |
| ... |
| 300.20171 |
| nan |
| 303.46467 |
| 321.71 |
| nan |
| nan |
| 332.05679 |
| 333.43715 |
| 342.47273 |
| nan |
| 350.72079 |
Let’s recap what we’ve done so far, then make some plots with the
data. Our data file has an extra line above the column names, so we
use the header_start keyword to tell it to start from line 1 instead
of line 0 (remember Python is 0-indexed!). We then used had to specify
that the data starts on line 2 using the data_start
keyword. Finally, we note some columns have missing values.
data = ascii.read("Young-Objects-Compilation.csv", header_start=1, data_start=2)
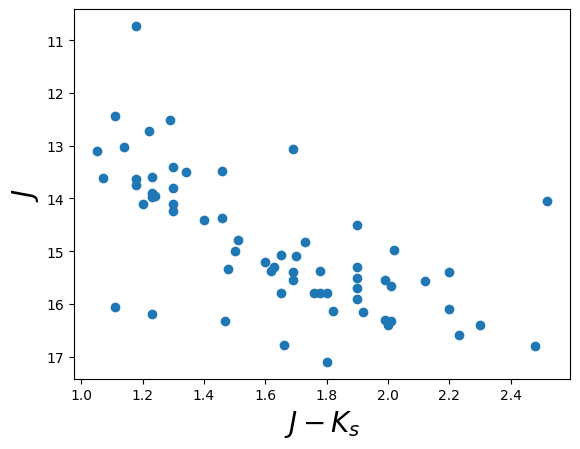
Now that we have our data loaded, let’s plot a color-magnitude diagram.
Here we simply make a scatter plot of the J-K color on the x-axis
against the J magnitude on the y-axis. We use a trick to flip the
y-axis plt.ylim(reversed(plt.ylim())). Called with no arguments,
plt.ylim() will return a tuple with the axis bounds,
e.g. (0,10). Calling the function with arguments will set the limits
of the axis, so we simply set the limits to be the reverse of whatever they
were before. Using this matplotlib-style plotting is convenient for
making quick plots and interactive use, but is not great if you need
more control over your figures.
plt.scatter(data["Jmag"] - data["Kmag"], data["Jmag"]) # plot J-K vs. J
plt.ylim(reversed(plt.ylim())) # flip the y-axis
plt.xlabel("$J-K_s$", fontsize=20)
plt.ylabel("$J$", fontsize=20)
Text(0, 0.5, '$J$')
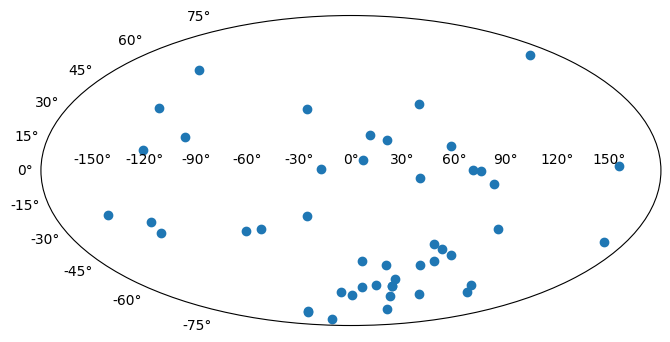
As a final example, we will plot the angular positions from the
catalog on a 2D projection of the sky. Instead of using matplotlib-style
plotting, we’ll take a more object-oriented approach. We’ll start by
creating a Figure object and adding a single subplot to the
figure. We can specify a projection with the projection keyword; in
this example we will use a Mollweide projection. Unfortunately, it is
highly non-trivial to make the matplotlib projection defined this way
follow the celestial convention of longitude/RA increasing to the left.
The axis object, ax, knows to expect angular coordinate
values. An important fact is that it expects the values to be in
radians, and it expects the azimuthal angle values to be between
(-180º,180º). This is (currently) not customizable, so we have to
coerce our RA data to conform to these rules! astropy provides a
coordinate class for handling angular values, astropy.coordinates.Angle.
We can convert our column of RA values to radians, and wrap the
angle bounds using this class.
ra = coord.Angle(data["RA"].filled(np.nan) * u.degree)
ra = ra.wrap_at(180 * u.degree)
dec = coord.Angle(data["Dec"].filled(np.nan) * u.degree)
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(111, projection="mollweide")
ax.scatter(ra.radian, dec.radian)
<matplotlib.collections.PathCollection at 0x7f97f8ffffe0>
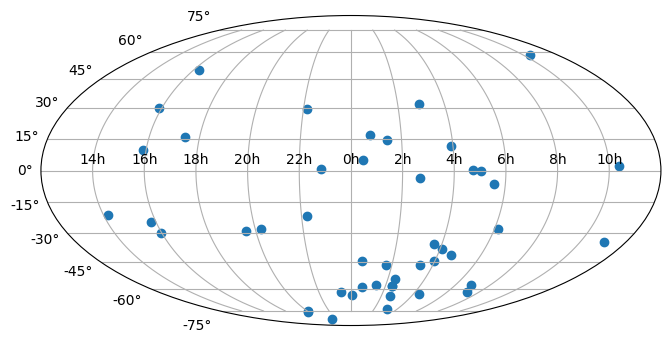
By default, matplotlib will add degree tick labels, so let’s change the horizontal (x) tick labels to be in units of hours, and display a grid:
fig = plt.figure(figsize=(8, 6))
ax = fig.add_subplot(111, projection="mollweide")
ax.scatter(ra.radian, dec.radian)
ax.set_xticklabels(
["14h", "16h", "18h", "20h", "22h", "0h", "2h", "4h", "6h", "8h", "10h"]
)
ax.grid(True)
We can save this figure as a PDF using the savefig function:
fig.savefig("map.pdf")
Exercises#
Make the map figures as just above, but color the points by the 'Kmag' column of the table.
Try making the maps again, but with each of the following projections: aitoff, hammer, lambert, and None (which is the same as not giving any projection). Do any of them make the data seem easier to understand?